Hohmann Lab
Research Publications Group Members BiographyEvolutionary play in gene expression
Regulation · Repurposing · Hijacking
Gene expression is fundamental to life, converting the information in DNA into proteins. Throughout evolution, cells have developed a rich variety of mechanisms to fine-tune and adapt gene expression to different environments and stresses.
When we think of eukaryotic gene expression, transcription in the nucleus and translation in the cytoplasm usually come to mind. Yet in between lies an essential, often overlooked step: nuclear mRNA export. This process ensures that only properly matured transcripts are delivered from the nucleus to the cytoplasm for translation. How are mRNAs distinguished from all other nuclear RNAs? How does the cell guarantee that only fully processed transcripts leave the nucleus? How are faulty mRNAs and transcription by-products targeted for degradation?
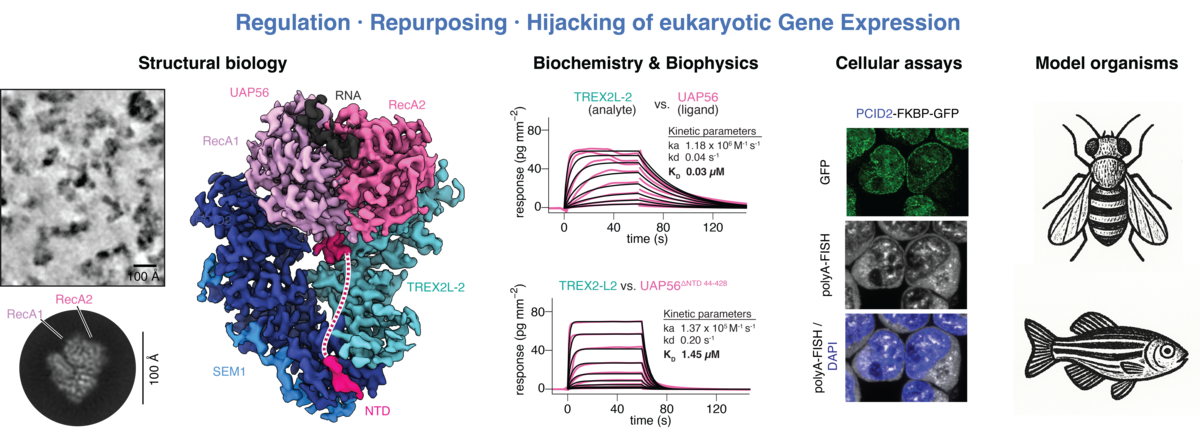
We are driven to discover new mechanisms in gene expression biology and to unveil their inner workings in mechanistic detail. Currently, we focus on three parallel entry points: (1) studying the regulation of mRNA export and its intersection with other nuclear RNA pathways, (2) understanding how mRNA export is hijacked in the animal germline for the processing of unusual RNAs, and (3) screening for novel intersections between viral proteins and host factors.
Mechanisms of mRNA export regulation
At the core of mRNA nuclear export lies an evolutionarily conserved machinery centred on the RNA clampase UAP56. We recently uncovered the mechanistic framework of the core of this pathway, where UAP56 orchestrates nuclear remodelling steps through its ATP-dependent RNA-binding cycle (for details see Hohmann et al, bioRxiv, 2024). However, the regulatory layers and connections to diverse cellular processes remain cryptic.
Intriguingly, we identified multiple novel direct interactors of UAP56, which we hypothesise to either stall or shortcut mRNA export. Using a combination of biochemistry, biophysics and integrative structural biology (including in silico predictions with AlphaFold and cryo-electron microscopy), we will reconstitute and characterise these new complexes in vitro and use cell culture systems to probe their functions in vivo. We are also interested in protein interactome mapping through proteomics and in CRISPR screens to uncover new interesting crosstalk between mRNA export and other cellular processes.
Repurposing of the mRNA export machinery in the germline
Gene expression in the animal germline faces unusual challenges, where canonical programmes are bypassed or deviated. This is true for both unusual RNAs, such as the piRNA precursor in the ovary of the fruit fly, which lacks all hallmark features of mRNAs and yet is exported from the nucleus by hijacking the mRNA export pathway, as well as for some mRNAs. For example, mRNAs in the testis are transcribed long before they are translated during sperm maturation. Making use of zebrafish and fruit flies as powerful genetic model systems, we want to uncover the mechanisms for how gene expression rules are bent in the germline. Besides our curiosity for unusual mechanisms, understanding these ‘quirks’ also holds the promise of uncovering previously unappreciated aspects in general gene expression biology.
Viral hijacking of the gene expression machinery
Viruses must rewire the host’s gene expression to replicate. Some block mRNA export to silence host defences, while others hijack export factors to favour viral RNAs. Historically, the virus-host arms race revealed many core principles of biology. We have already discovered previously uncharacterised viral interactions with the export machinery and are curious to understand their function. We are interested in building on our previous knowledge for the clever use of established screens and the development of new large-scale screens to uncover new interactions of the host gene expression machinery with viral factors.
Approaches
We combine the power of both in vitro and in vivo approaches to discover mechanisms in gene expression biology. Our strengths are biochemistry, where we reconstitute protein-RNA complexes for biophysical analysis, as well as structural biology, where we employ in silico modelling approaches such as AlphaFold and cryo-electron microscopy. We complement these in vitro approaches with work in cell culture systems, as well as zebrafish and the fruit fly as powerful genetic model organisms to test and expand our hypotheses in vivo. Here we use a wide array of methods ranging from genome editing using CRISPR/Cas, light microscopy and reporter design to proteomics and transcriptomics.
We believe that science is best done, and much more fun, in a collaborative setting. In our lab, we value both independence and teamwork, and enjoy collaborating with other groups.
Recruitment
Most importantly, we are driven by curiosity and the joy of discovery in a curious, creative, collaborative and supportive environment. If you can relate to the child-like excitement that scientific ‘aha moments’ can cause, please get in touch!