Fluorescent timers reveal the secrets of protein degradation
RESEARCH HIGHLIGHT
28 June- The research team of Anton Khmelinskii at the Institute of Molecular Biology (IMB) published a paper this month in Molecular Cell, in which they use a fluorescent timer system to systematically characterise the protein degradation system of yeast cells. In this study, they identify the substrates of most of the protein players in the degradation system and identify a new receptor that is required for substrate recognition. This creates an extremely useful information resource and opens new ways to study protein degradation on a massive scale.
In order to function properly, cells must be able to express all of the correct proteins in the correct amounts. Conversely, any unnecessary or abnormal proteins must be removed, or degraded, to prevent them from impairing cell function, as this can lead to diseases such as cancer and neurodegenerative disorders.
In eukaryotes, protein degradation is largely performed by a network of enzymes called the ubiquitin-proteasome system (UPS). In this system, proteins that should be degraded are marked with an ubiquitin label by an enzymatic cascade involving ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin ligase (E3) enzymes. However, the UPS is vast – humans have more than 600 E3s and yeast about 100 – and the functions of many enzymes in the UPS remain unclear.
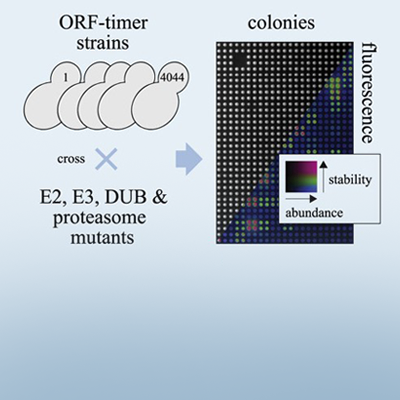
In this study, Anton’s group sought to gain a better understanding of the UPS in budding yeast. Key to this was a technique that he had previously developed with tandem fluorescent protein timers (tFTs). This method works by tagging proteins of interest with two fluorescent markers, one red (mCherry) and one green (superfolder GFP). The superfolder GFP tag matures faster than the mCherry tag, such that green fluorescence reflects protein abundance, while the ratio of red:green fluorescence reflects protein stability, or how quickly the protein is degraded. By monitoring the amount of green and red fluorescence, abundance and turnover of any tagged protein can be assessed.
Anton had previously created a library of 4,044 yeast strains in which each protein in the yeast proteome is tagged with a tFT tag. Now with collaborators at the University of Heidelberg, EMBL and Toronto, the team crossed each of these library strains to an array of 132 yeast strains, each with a mutation in a different UPS component. This was a massive effort, involving ∼2.5 million crosses and more than 620,000 UPS mutant-tFT strains. But it created a highly useful resource that allowed them to systematically determine the effect of each UPS component on the abundance and stability of each protein in the proteome.
The result was a trove of data, which Anton and his colleagues used to make discoveries about many different UPS components. One of the most important findings they made was identifying a new receptor in an E3 called the glucose-induced degradation-deficient (GID) complex, which orchestrates protein degradation in response to changes in nutrition. Edwin (the first author of this study) noticed that this new receptor, which they named Gid11, mostly recognised proteins with an N-terminal threonine. When Edwin mutated the N-terminal threonine, the abundance and stability of these proteins was no longer regulated by Gid11. He also showed that Gid11 expression was induced in cells metabolising ethanol, a non-fermentable carbon source, and in response to different stresses. This highlights that Gid11 is a conditionally expressed GID receptor that degrades proteins with an N-terminal threonine.
Overall, this study creates a rich dataset that other researchers can use to explore different functions of the UPS. Moreover, it demonstrates that the tFT assay is a highly useful tool that can open new ways for researchers to explore protein degradation on a large scale. In future, Anton and his lab hope to further characterise the precise N-terminal motifs recognised by Gid11 and identify more GID substrate receptors to gain a more complete understanding of this enigmatic ubiquitin ligase.
Cheryl Li is a Science Writer at the Institute of Molecular Biology (IMB)
Further details
Further information can be found at https://www.sciencedirect.com/science/article/pii/S1097276521003233
Anton is a Group Leader at IMB. Further information about research in the Khmelinskii lab can be found at www.imb.de/khmelinskii.
About the Institute of Molecular Biology gGmbH
The Institute of Molecular Biology gGmbH (IMB) is a centre of excellence in the life sciences that was established in 2011 on the campus of Johannes Gutenberg University Mainz (JGU). Research at IMB focuses on three cutting-edge areas: epigenetics, developmental biology, and genome stability. The institute is a prime example of successful collaboration between a private foundation and government: The Boehringer Ingelheim Foundation has committed 154 million euros to be disbursed from 2009 until 2027 to cover the operating costs of research at IMB. The State of Rhineland-Palatinate has provided approximately 50 million euros for the construction of a state-of-the-art building and is giving a further 52 million in core funding from 2020 until 2027. For more information about IMB, please visit: www.imb.de.
Boehringer Ingelheim Foundation
The Boehringer Ingelheim Foundation is an independent, non-profit organization that is committed to the promotion of the medical, biological, chemical, and pharmaceutical sciences. It was established in 1977 by Hubertus Liebrecht (1931–1991), a member of the shareholder family of the Boehringer Ingelheim company. Through its Perspectives Programme Plus 3 and its Exploration Grants, the Foundation supports independent junior group leaders. It also endows the international Heinrich Wieland Prize, as well as awards for up-and-coming scientists in Germany. In addition, the Foundation funds institutional projects in Germany, such as the Institute of Molecular Biology (IMB), the department of life sciences at the University of Mainz, and the European Molecular Biology Laboratory (EMBL) in Heidelberg.
Press contact for further information
Dr Ralf Dahm, Director of Scientific Management
Institute of Molecular Biology gGmbH (IMB), Ackermannweg 4, 55128 Mainz, Germany
Phone: +49 (0) 6131 39 21455, Fax: +49 (0) 6131 39 21421, Email: press(at)imb.de