Roukos Lab
Research Publications Group Members BiographyChromosome fragility & 3D chromosome organisation
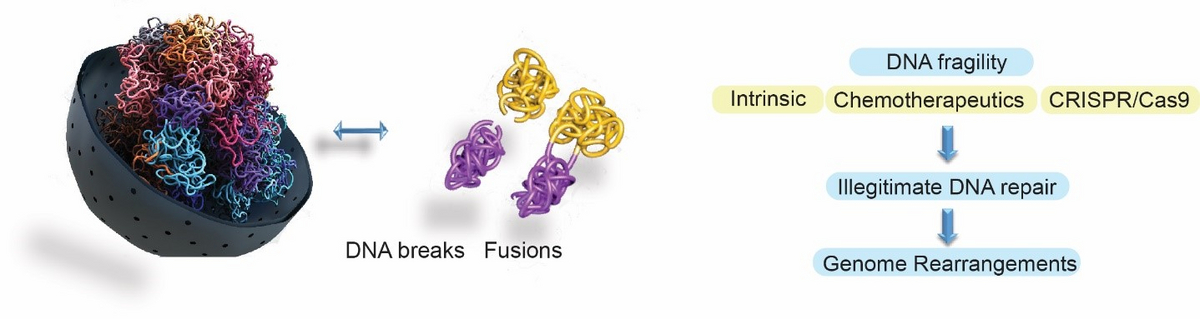
Our lab focuses on understanding how cells maintain the stability of their genome in the context of 3D genome organisation. We are interested in shedding light on when, where and why chromosomes become fragile, how this mechanistically occurs, and how DNA repair mechanisms prevent the illegitimate joining that leads to the formation of oncogenic genome rearrangements. Our work is focused on intrinsic or exogenous sources of DNA fragility, or on DSBs that form upon the use of CRISPR/Cas nucleases. A main goal of our research focuses on understanding why treatment with widely-used chemotherapeutics leads to the formation of chromosome fusions that drive secondary leukaemias. Another pillar of our research focuses on how perturbation of intrinsic nuclear processes, such as transcription, replication and chromatin loop extrusion, promotes endogenous DNA damage in various tissues and stem cells of the human body, and how this is linked with the formation of cell type- and tissue-specific oncogenic genome rearrangements. Finally, our goal is to shed light on the molecular determinants of off-target activity and the scission profile of Cas nucleases in order to minimise unwanted off-target genomic instability and establish frameworks for predicted and precise genome editing. To address our research questions, we develop and use a combination of state-of-the-art high-throughput imaging and next-generation sequencing methodologies.
DNA topology, chromosome organisation & genomic instability
Spatial chromosome architecture is a key factor in eukaryotic gene regulation and genome stability, but how changes in local DNA topology coordinate with global 3D chromosome organisation remains poorly understood. Type II topoisomerases (TOP2s) resolve torsional stress accumulated during various cellular processes and are enriched at chromatin loop anchors and TAD boundaries, where, when trapped, as shown recently by our lab and others, can lead to genomic instability promoting the formation of oncogenic fusions (Gothe et al., 2019, Mol Cell). However, whether TOP2s relieve topological constraints associated with chromatin loop extrusion at these positions and/or participate in 3D chromosome folding remains unclear. Our lab is particularly interested in understanding how topoisomerases control DNA topology and chromosome organisation by remodelling DNA supercoiling accumulating during cellular processes such as replication and transcription. We are also interested in understanding how perturbation of the coordination of DNA topology and chromosome organisation can lead to genomic instability, which can drive cancer and other disorders.
Sources and consequences of intrinsic genomic instability across human cell types
In this project funded by the SFB 1361, we aim to uncover how the influences of transcription, DNA topology and replication converge to determine which regions of the genome preferentially break. We investigate whether intrinsic DNA fragility varies between different human cell types and stem cells, and how this relates to genomic rearrangements found in cancer. To address this, we include a fully-fledged bioinformatics pillar in collaboration with Kathi Zarnack’s lab to combine elaborate genomics approaches with multi-modal data integration, quantitative comparisons and machine learning. Our project thereby provides both experimental and computational tools to study genomic instability and strongly contributes to the enhanced emphasis of the consortium on systems approaches.
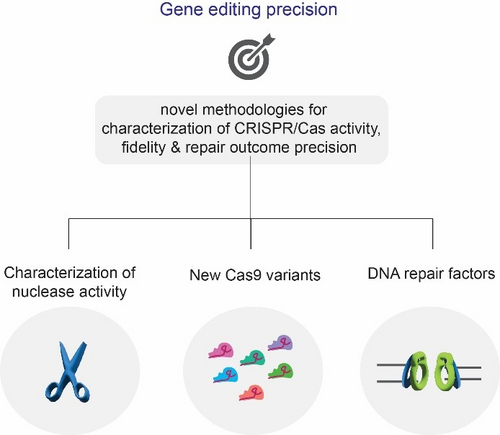
Genome editing precision
CRISPR/Cas9 is a revolutionary genome engineering platform with the potential to correct mutations responsible for several diseases, but off-target effects and gene editing precision can hinder the success of gene editing therapy. In our lab, we investigate pathways that can contribute to controlled genome editing using CRISPR/Cas. To this end, we have recently developed a novel NGS-based method for the characterisation of the CRSPR/Cas9 off-target landscape and identifying determinants of the Cas9 nuclease cleavage profile (Longo, Sayols et al., Nat Biotech, 2024, in press). We show that by controlling the DNA double-strand break (DSB) end structure of Cas9 cuts, we can increase gene editing precision. Moreover, we characterise the scission profile of high-fidelity Cas9 variants for better control of Cas9-induced mutations and demonstrate that the cleavage patterns can be used for correcting pathogenic deletions. Building upon this knowledge, we aim to generate novel Cas9 variants with higher cleavage profile flexibility and manipulate DNA repair pathways to ultimately dictate genome editing outcomes.
Our group has recently relocated to the Medical School, University of Patras, Greece, but we still keep an affiliation at IMB.
We are constantly looking for highly motivated researchers who enjoy working in a multi-disciplinary and collaborative environment to join our group.