Niehrs Lab
Research Publications Group Members BiographyDNA demethylation, DNA repair & reprogramming
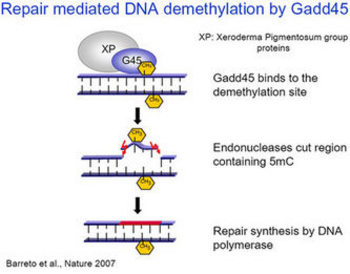
Epigenetic gene regulation is of central importance for development and disease. In the genome of many multicellular organisms, DNA methylation is a common epigenetic mark associated with gene silencing. DNA methylation plays important roles in gene expression, genomic imprinting, X-chromosome inactivation, genomic instability, embryonic development and cancer. The pattern of DNA methylation varies in different tissues and during embryonic development and little is known how this methylation pattern is regulated. DNA methylation is a dynamic process and can be reversed by enzymatic demethylation involving TET cytosine dioxygenases, a process that is still incompletely understood. DNA demethylation is a widespread phenomenon, occurring in plants as well as in animals during development, and in the adult. For example, somatic cell reprogramming is proceeded by the reactivation of pluripotency genes. Part of this reactivation is DNA demethylation. We showed that Growth arrest and DNA damage 45A (GADD45A ) is a key player in active DNA demethylation and acts via DNA repair (Figure 1). One goal of our research is to analyse the mechanism of DNA demethylation as well as the role played by GADD45A in development. Our results indicate that GADD45A acts as an adapter protein, which directs DNA methylation machinery to specific loci. To address GADD45A function, we use biochemical, molecular biology and cell biological approaches, employing the mouse and frog model systems as well as embryonic stem cells.
Role of noncoding RNA in demethylation
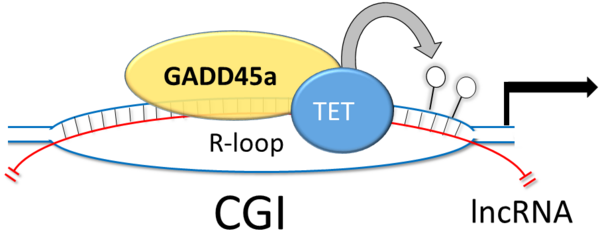
A key question in DNA demethylation is how the process is targeted to specific loci. To be directed to specific genomic loci, GADD45A relies on bridging factors. We showed that long noncoding RNAs (lncRNAs) can serve as address labels for GADD45A to guide the demethylation machinery (Figure 2). This involves R-loops, DNA:RNA hybrids, forming at the target genes, which are bound by GADD45 to trigger local DNA demethylation. Conceptually, R-loops are attractive candidates because analogous to bacterial guide RNAs, which form R-loops to direct CAS9 endonuclease, mammalian R-loops may direct epigenetic modifiers in a sequence-specific manner to genomic loci. Our study indicates that GADD45A is an epigenetic reader of such regulatory R-loops at CGIs (Figure 2).
The goal of our research is to analyse the mechanism of DNA demethylation as well as the role played by GADD45A in development. Which target genes are regulated by GADD45A ? What are the sequence determinants that recruit GADD45A to specific genes during demethylation? What is the role of GADD45A -mediated demethylation in mammals?
To investigate these questions, we analyse mouse embryonic stem cells (mESC) and transgenic mice that are mutant for GADD45. Applying genome-wide approaches and studying the biology of ESCs, we will systematically identify and characterise regulatory R-loops recognised by GADD45, to address how R-loops are decoded via demethylation in the context of ESC pluripotency and differentiation, as well as during mouse development.
DNA demethylation & ageing
Changes in DNA methylation are among the best-documented epigenetic alterations, which accompany ageing. However, if and how altered DNA methylation is causally involved in ageing has remained elusive. We found that Gadd45a/Ing1 double knockout mice (DKO) display premature ageing and phenocopy impaired energy homeostasis and lipodystrophy characteristic of Cebp (CCAAT/enhancer binding protein) mutants. Correspondingly, GADD45α occupies C/EBPβ/δ- dependent super-enhancers, and cooperatively with ING1 promotes local DNA demethylation to permit C/EBPb recruitment. This indicates that enhancer methylation affects ageing. We now analyse the role of CEBP and demethylation in ageing models, as this may lead to novel avenues to promote healthy ageing.