Lemke Lab
Research Publications Group Members BiographySynthetic biophysics of protein disorder
We focus on studying intrinsically disordered proteins (IDPs), which constitute up to 50% of the eukaryotic proteome. Paradoxically, they are most famous for their involvement in misfolding in neurodegenerative diseases of ageing like Alzheimer, Parkinson and Huntington disease. However, IDPs are key in many vital biological processes, such as nucleocytoplasmic transport, transcription and gene regulation. The ability of IDPs to exist in multiple conformations is considered a major driving force behind their enrichment during evolution in eukaryotes. Studying biological machineries containing such dynamic proteins is a major hurdle for conventional technologies. Because of this, and due to the fact that they are hard to visualise, IDPs are termed the dark proteome. Using a question-driven, multidisciplinary approach paired with novel tool development, we have made major strides in understanding the biological dynamics of such systems from the single molecule to the whole cell level. Fluorescence tools are ideally suited to study the plasticity of IDPs, since their non-invasive character permits smooth transition between in vitro (biochemical) and in vivo (in cell) studies. In particular, single molecule and super-resolution techniques are powerful tools for studying the spatial and temporal heterogeneities that are intrinsic to complex biological systems. We synergistically combine this effort with cutting-edge developments in chemical and synthetic biology, microfluidics and microscope engineering to increase the throughput, strength and sensitivity of the approach as a whole.
Research highlights
Our strong focus on understanding the mechanisms of IDP function is both driven by and driving novel tool developments for “in-cell/in situ structural biology.” This comprises a synergistic effort of chemical/synthetic biology and precision fluorescence-based technology/nanoscopy/single-molecule/super-resolution/microfluidics development.
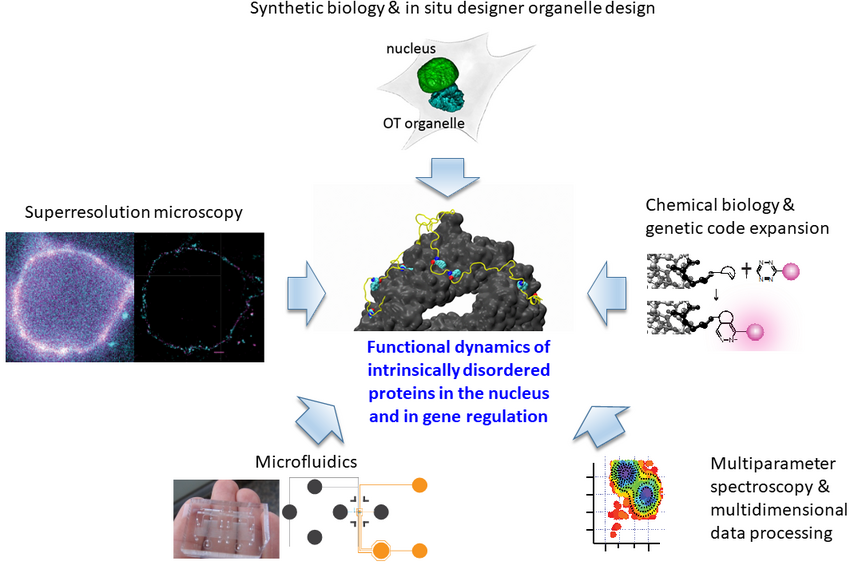
A major technical breakthrough of my lab was the ability to engineer “click”able functionalities into any protein in vitro and in vivo. This genetic code expansion (GCE) approach has the potential to become a true GFP (fusion protein) surrogate strategy, with the major advantages being that superior synthetic dyes can be coupled with residue-specific precision anywhere in a protein. This opens up new avenues in single-molecule fluorescence and super-resolution microscopy. More recently, we have been able to merge our understanding of protein disorder and synthetic biology into the design of new membraneless organelles dedicated to protein engineering in situ (Figure 1). These custom organelles do not just execute a distinct second genetic code inside the cells; their bottom-up design also enables us to learn how phase separation can be used to generate new functions in eukaryotes. Our findings also have wide implications for understanding gene regulatory and stress-based mechanisms carried out by distinct, naturally occurring organelles that play vital roles in regular cell function, as well as in ageing mechanisms.
These precision tools enable us to make even the most complex molecular machinery visible to our core methodologies, which are based on time-resolved multiparameter and nanoscopy tools. This allows innovative approaches to study the heterogeneity of IDPs. We discovered a distinct ultrafast protein-protein interaction mechanism that can explain how nuclear pore complexes (NPCs) efficiently fulfil their central role in cellular logistics, and how nuclear transport can be both fast and selective at the same time. We also used microfluidics to show how the permeability barrier of the nuclear envelope could be formed by liquid-liquid phase separation of a single disordered protein species. These findings provide a leap forward in our understanding of how IDPs maintain different functionalities through conformational changes, despite the normal requirement for rigid molecular specificity.
Future directions
IDPs lack a stable structure and can easily misfold to the amyloid state and aggregate, leading to their prominent role in many ageing-related diseases. This intrinsic risk must be outweighed by multiple advantages to explain their enrichment in evolution of more complex species, but we are only at the beginning of understanding this. IDPs are highly multifunctional, and due to their multivalency and large, disordered regions they can function as dynamic scaffold platforms. We combine chemical and synthetic biology approaches to enable non-invasive, multi-colour high- and super-resolution studies of activity-dependent protein conformation changes in living cells, enabling fluorescence-driven in situ structural biology. The key point is that the enhanced spatial and temporal resolution offered by our fluorescent methods will enable us to detect rare events and unexpected behaviours inside cells. We use this to identify and understand IDP multifunctionalities that are clearly distinguishable from their normal mode of action. For example, nucleoporins (Nups) normally function in the nuclear pore complex (NPC), but in fact many IDP-Nups have diverse roles, such as in pathogen-host interactions, and can even shuttle away from the NPC to function in gene regulatory processes. Fusions of Nup98 with transcription factors, for example, are known to be linked to leukaemia.
Our work is accompanied by rigorous analysis of the physicochemical properties of IDPs and examines to what extent simple, known polymer concepts such as phase separation can be used to describe the function of IDP biopolymers in vivo. Vice versa, we are particularly interested in how disordered proteins play key roles in gene regulation and cellular ageing.
For more about the Lemke lab click here for his personal lab webpage or here for his page at EMBL