Khmelinskii Lab
Research Publications Group Members BiographyProteome organisation & dynamics
The functional state of a cell is ultimately defined by the state of its proteome, i.e. abundance, localisation, turnover and mobility of all proteins and their organisation in complexes and organelles. Numerous cellular systems contribute to proteome homeostasis through prevention, detection and removal of abnormal proteins. These quality control systems operate throughout the protein life cycle, from synthesis to degradation. Hence the definition of abnormal protein is very broad, including mutated, misfolded, mislocalised and damaged molecules.
Selective protein degradation by the ubiquitin-proteasome system (UPS) plays a key role in proteome abundance and quality control, whereby a cascade of ubiquitin-activating (E1), ubiquitin-conjugating (E2) and ubiquitin-protein ligase (E3) enzymes marks proteins with polyubiquitin chains of varying linkages for degradation at the proteasome. When degradation is not possible, the impact of abnormal proteins can be mitigated through their asymmetric partitioning during cell division. Despite the activity of such systems, proteome homeostasis declines with ageing and in numerous diseases, including some cancers and late-onset neurodegenerative disorders, resulting in accumulation of abnormal proteins and loss of cell functionality. Thus, understanding the mechanisms of protein quality control is not only important from the point of view of basic research but can also contribute to elucidating causes of diseases at the molecular level.
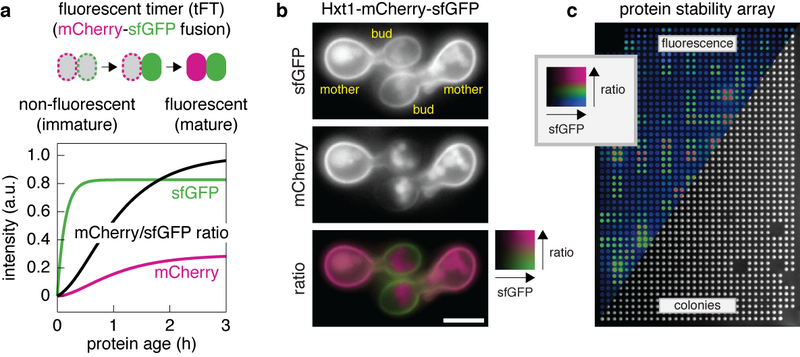
We employ a multidisciplinary approach to dissect mechanisms of protein quality control in yeast and human cells through a combination of molecular and cell biology techniques, biochemistry, computational biology and development of genomic and proteomic approaches (Figure 1) that allow us to follow proteome dynamics down to single cell level. Through systematic characterisation of selective protein degradation machinery and patterns of protein turnover/inheritance, we aim:
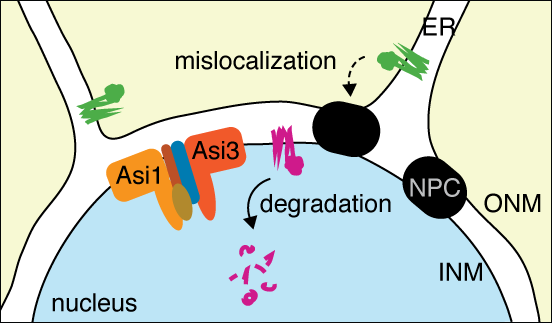
- to identify quality control mechanisms for different types of abnormal proteins (e.g. our recent work revealed a quality control pathway for proteins mislocalised to the inner nuclear membrane, Figure 2)
- to understand how abnormal proteins are recognised in the cell
- to examine the roles of protein quality systems in healthy cells, to investigate their contributions to disease progression and ageing, and to ultimately explore their potential as targets for therapeutic interventions
Recruiting
We are looking for highly motivated postdoctoral fellows, PhD students and Master students to join our group.
Postdoc applicants are encouraged to visit the IMB postdoc programme or contact Anton Khmelinskii directly by email, including a CV, research interests and/or project idea.
PhD applicants please apply through the International PhD Programme. See here for further details.
We welcome Master students or students interested an internship of at least 6 months. Please send your CV and a brief motivation letter to Anton Khmelinskii by email.