Ketting Lab
Research Publications Group Members BiographyBiology of non-coding RNA
Many RNAs in the cell do not code for proteins but have gene regulatory functions. We study non-coding RNA molecules, with a particular focus on a family of small RNA molecules that act in so-called RNA interference (RNAi)-related pathways. Since the discovery of RNAi-related pathways at the start of the 21st century, it has been evident that each pathway has its own unique characteristics and effects on gene expression. Nevertheless, they all share one common feature: a member of the Argonaute protein family that binds a cofactor in the form of a small RNA molecule. These small non-coding RNAs provide sequence specificity to the Argonaute proteins, and it is therefore clear that the identity of these small RNA molecules is critical for the functionality of gene regulatory pathways.
We focus on small RNA-driven gene regulatory mechanisms that act in the germline. These mechanisms play an important role in resolving genetic conflicts arising from the presence of parasites in our genomes: transposable elements. In order to control the activity of these potentially harmful elements, small RNAs that recognise these elements need to be made, while genes need to be expressed and not targeted. Studying how these small RNAs act and how their specificity to transposable elements can be achieved is a core interest of our group. In addition, we have a strong interest in how small RNA pathways are organised in phase-separated structures known as germ granules. We believe that these pathways represent excellent models to enhance our understanding of how phase separation functions inside cells in general and how it can be regulated. For instance, deregulated phase separation plays a role in the formation of aggregates in neurons, leading to neurodegenerative diseases. In ageing systems, such deregulated aggregates also play an important role, and a better understanding of how these structures function in healthy systems is badly needed to counter their effects. Finally, as these pathways are an integral part of the germ cell gene regulatory network, germ cell biology is also a key topic of interest in our lab. In particular, we have a keen interest in understanding the immortality of the germline. Every generation, a fresh and rejuvenated germline develops; hence this cell type appears to evade the ageing process that inevitably affects somatic cells. We mainly use two model systems for our studies: the nematode Caenorhabditis elegans and the zebrafish Danio rerio.
Germ granules & phase separation
Germ granules have long been known to be characteristic entities of germ cells. We now know that these granules represent membrane-less organelles formed by processes related to liquid-liquid phase separation (LLPS) and that different types of germ granules exist. It is also well-established that small RNA pathway components are often found in such germ granules, but the relevance of this sub-cellular localisation is still poorly understood. We are studying how and why Argonaute proteins and other RNAs and proteins localise to different types of germ granules and how this may change with time as a function of, for instance, germ cell development or Argonaute protein maturation. These studies aim to better understand small RNA function and germ cell development and to provide valuable fundamental insights into how LLPS functions and is regulated in cells. As such, we see germ cells as a rich and versatile system to study many different aspects of LLPS in biological systems.
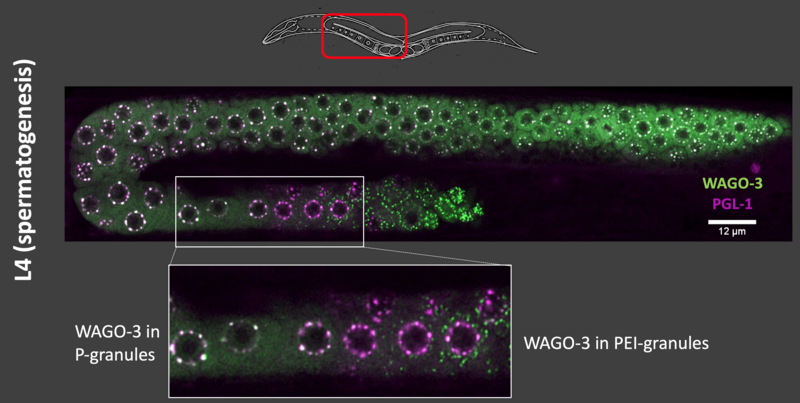
A fascinating aspect of small RNAs in germ cells is that they can be transmitted into the next generation by being specifically retained in mature oocytes and sperm. This is key for maintaining the small RNA populations that are effective in controlling transposable elements, and at the same time does not interfere too much with normal gene regulation. However, as small RNAs are also involved in the regulation of ‘normal’ (non-TE-related) genes, this phenomenon of inheritance also provides opportunities to transmit gene regulatory information across generations. Recently, we identified a novel type of germ granule - the PEI granule (Figure 2). These granules are required for the inheritance of loaded Argonaute proteins via the sperm. In the context of the CRC 1551 research network, we are currently following up on our published work by studying how PEI granules form and function using a combination of biochemical, modelling and genetic approaches.
Understanding small RNA biogenesis
In order to function as a genomic immune system, small RNAs must be able to recognise a vast and continuously changing repertoire of sequences. How can cells achieve such a formidable task, while at the same time avoid silencing genes that should be expressed? To understand this, we need to understand the mechanisms that generate small RNAs. Interestingly, small RNA pathways with such immune-like functions have a diverse repertoire of small RNA biogenesis mechanisms. This is in stark contrast to miRNAs, which have evolutionarily well-conserved biogenesis pathways. We believe that this relates to an ongoing arms race between transposons and small RNA-driven pathways in the germline, driving fast evolution of these mechanisms. Because of this, we study the process of small RNA biogenesis. In one of our projects, which is part of the campus-wide graduate programme GenEvo, we study how an evolutionarily well-conserved protein has adapted to diverse roles in different nematodes, including C. elegans. We additionally have a number of projects that aim to identify and characterise novel proteins in the biogenesis process. Notably, we also have an interest in understanding how small RNA-generating loci are transcribed. We are also interested in how these fast-evolving mechanisms are interwoven into the many other core gene regulatory mechanisms in the cell, including the possible involvement of RNA modifications. Both genetic and biochemical approaches are used in these experiments.
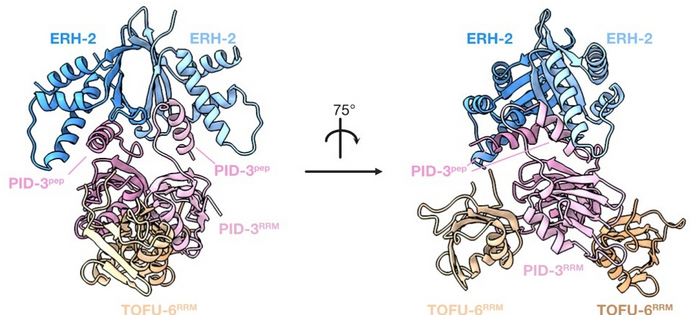
Approaches
We use C. elegans and zebrafish to study these processes by genetics and microscopy. We employ CRISPR-Cas genome editing, mutagenesis and transgenesis techniques daily, as well as confocal, spinning disk and super-resolution microscopy. We also use bioinformatics to analyse small RNA and other types of nucleic acid sequencing data. Finally, we are using biochemical approaches to further refine our ideas on how the identified factors operate together.
Recruitment
We are an international, highly diverse and competitive team full of enthusiastic people who love science. Importantly, we also appreciate a healthy work-life balance. We are always looking for motivated new colleagues! If you have an interest in studying germ granules, small RNA biology, genetics, microscopy, biochemistry, bioinformatics, or any combination thereof, maybe you can be our next recruitment! Please send us an e-mail if we raised your interest.