Superresolution Microscopy of Functional Nuclear Nanostructure
Functional nuclear organization has emerged as an important topic of epigenetics. For this, methods of far field light optical resolution are required beyond the possibilities of conventional epifluorescence microscopy (optical resolution about 200 nm laterally, 600 nm axially). Towards this goal, we have established a variety of superresolution microscopy (“nanoscopy”) methods.
Our present spectrum for 'nanoimaging' of nuclear structures comprises confocal laser scanning 4Pi-microscopy, Spatially Modulated Illumination (SMI) and Patterned Excitation Microscopy (PEM) devices, and Spectrally Assigned Localization Microscopy (SALM).
While 4Pi-microcoscopy was applied to the superresolution of nuclear pore complex distribution, replication complexes and other nuclear nanostructures (axial optical resolution in the 120 nm range), SMI microscopy made it possible to measure the size of telomeric complexes with a resolution down to a few tens of nanometer and to perform precise size measurements of the compaction status of small, specifically labelled, chromatin domains.
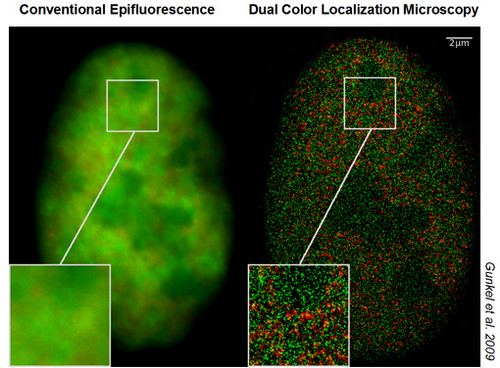
Using a recently developed SALM technique, Spectral Precision Distance/Position Determination Microscopy (SPDM) with Physically Modifiable Fluorophores (SPDMPhymod), nuclear nanostructures can now be studied on a large scale in 3D intact nuclei down to a lateral optical resolution of individual molecules in the macromolecular range in optical sections of ca. 600 nm thickness. This technique can be performed with standard fluorescence proteins/fluorochromes and is applicable also in vivo. For example, the distribution of individually resolved nuclear pore complex proteins, histones, or FISH labelled repetitive short DNA sequences was determined with a lateral optical resolution down to about 15 nm; the spatial location of two species of single molecules in human cell nuclei (e.g. histones and chromatin remodelling factors; histones and polymerase II) was determined simultaneously by dual color localization microscopy up to a density of ca. 1000 molecules/µm². Nanoscopy experiments of other cellular features combining SPDMPhymod and Structured Illumination Microscopy (SIM) indicate that in this way, appropriately labeled chromatin structures may be analysed in 3D intact cells at an optical 3D resolution of 40-50 nm.
These superresolution microscopy methods can also be applied to other biological nanostructures. Our present experience using SPDM comprises single molecule resolution in membranes, cell junctions, bacteria, and viruses. Under optimum conditions, we presently achieve an optical resolution potential of 5 nm (ca. 1/100 of the exciting wavelength).
Perspectives
The focus of our future research at IMB will be to further improve these methods and apply them in collaborative projects in epigenetics.
In addition, our scientists develop super-resolution light microscopes to probe the functional nanostructure of the nucleus. These microscopes can resolve structures beyond the diffraction limit, allowing the visualization of biological structures in unprecedented detail.
Furthermore, the Cremer group hosts a range of training events. Details of these events can be found here.
eXtreme Resolution Microscopy (XMR)
For further information regarding the Cremer group, please see their eXtreme Resolution Microscopy (XMR) website found here.